Free phylogenetic tree creator
Unveil evolutionary history using our free, powerful phylogenetic tree maker.
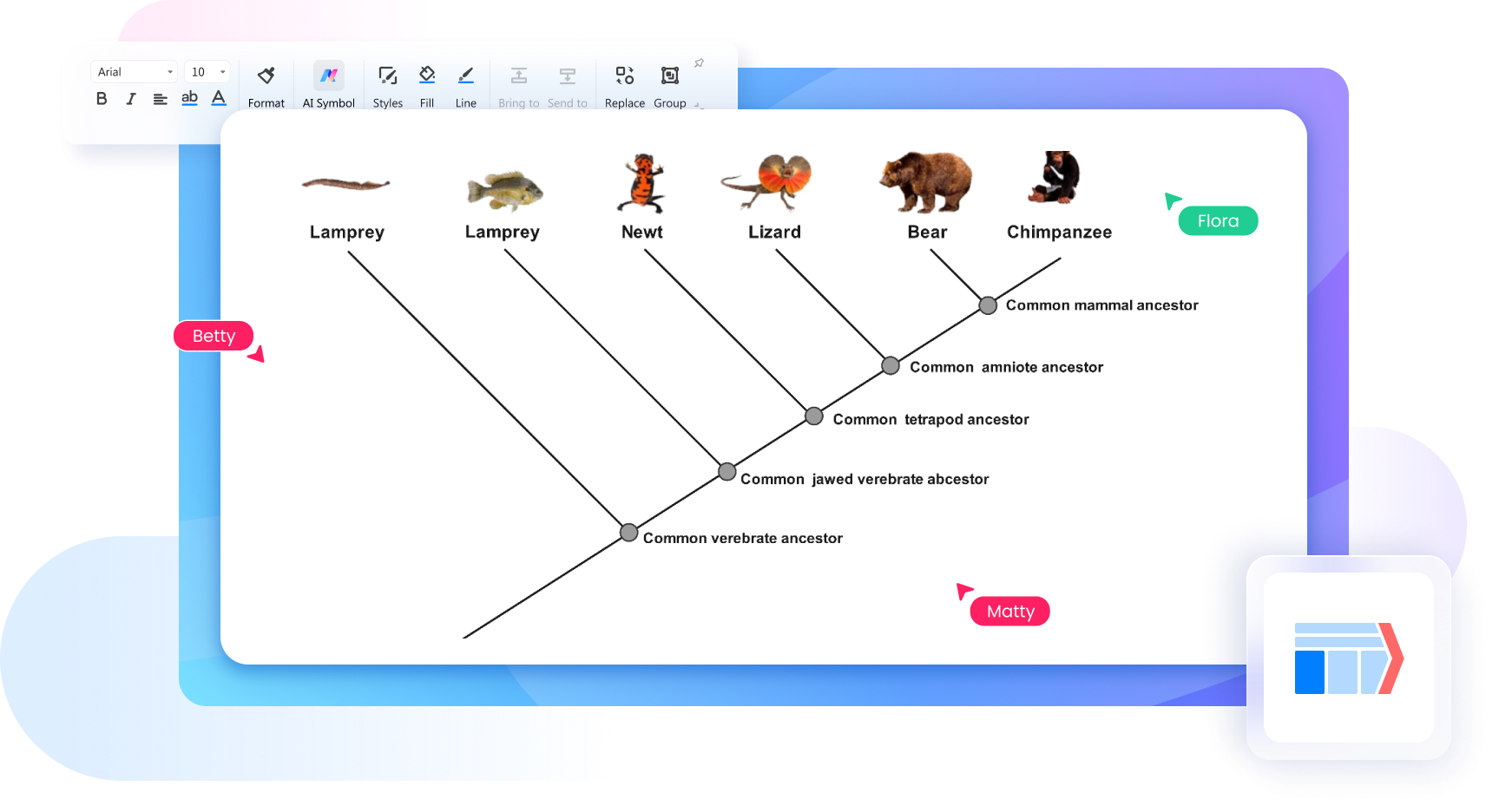

Your easy-to-user phylogenetic tree maker
Easy to create
Create your phylogenetic tree diagrams effortlessly. Adjust layouts, labels, and styles for personalized research representation. Use EdrawMax’s free symbols, icons, cliparts and various other elements for seamless evolutionary connections.
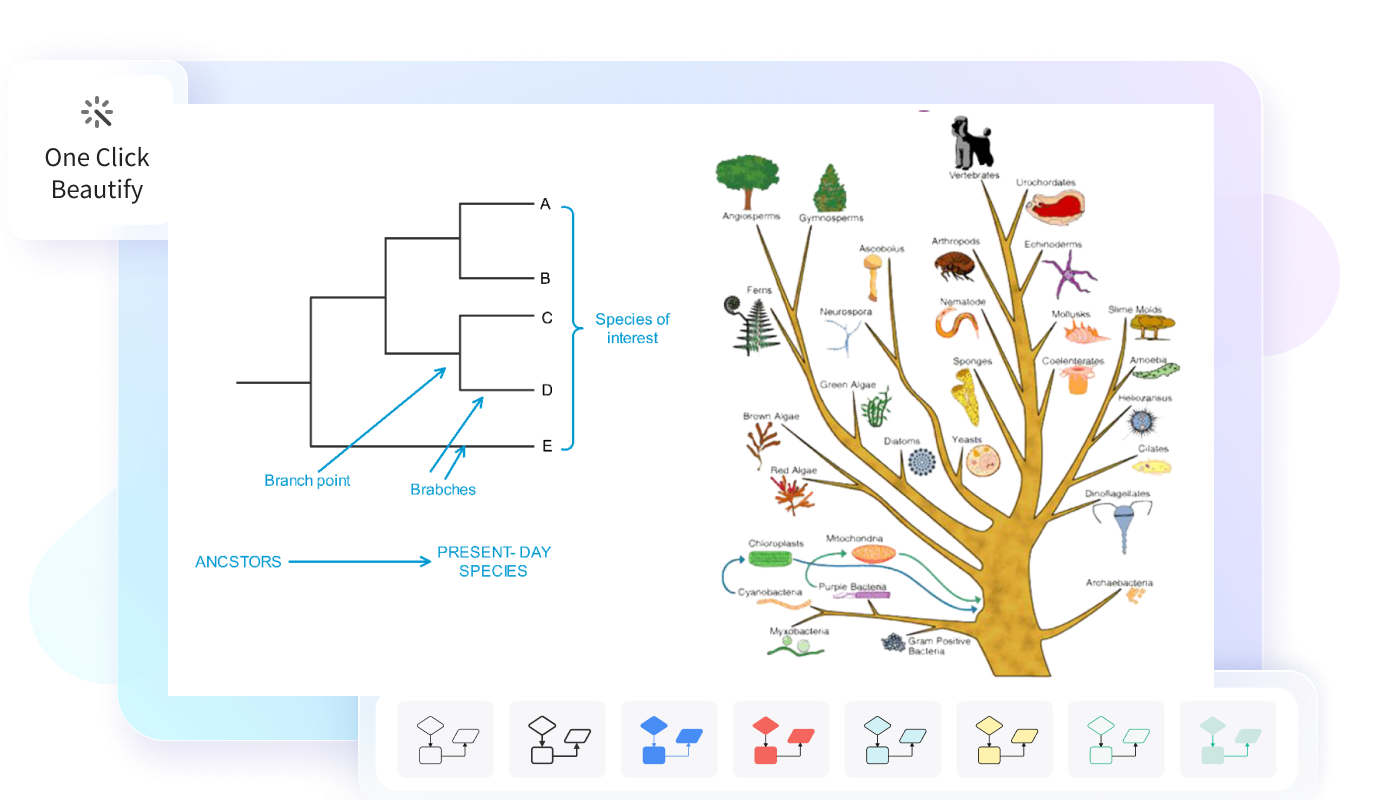
Rich templates
EdrawMax offers numerous phylogenetic tree templates and 16,000+ all kinds of templates, which could also be helpful. With these elaborately made templates, you can effortlessly get started.
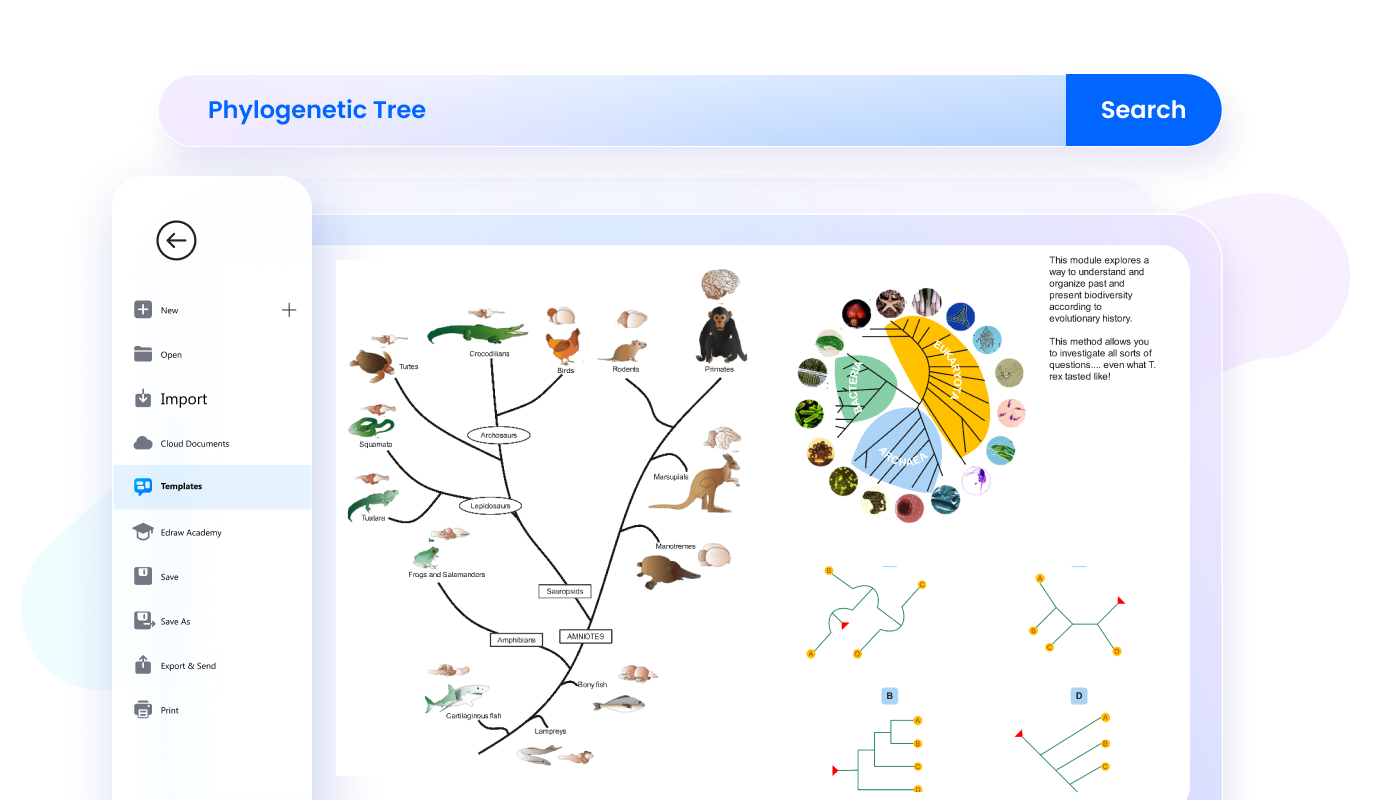
Seamless teamwork
Store phylogenic trees on cloud and grant access to specific team members, allowing them to view and provide feedback on your work.
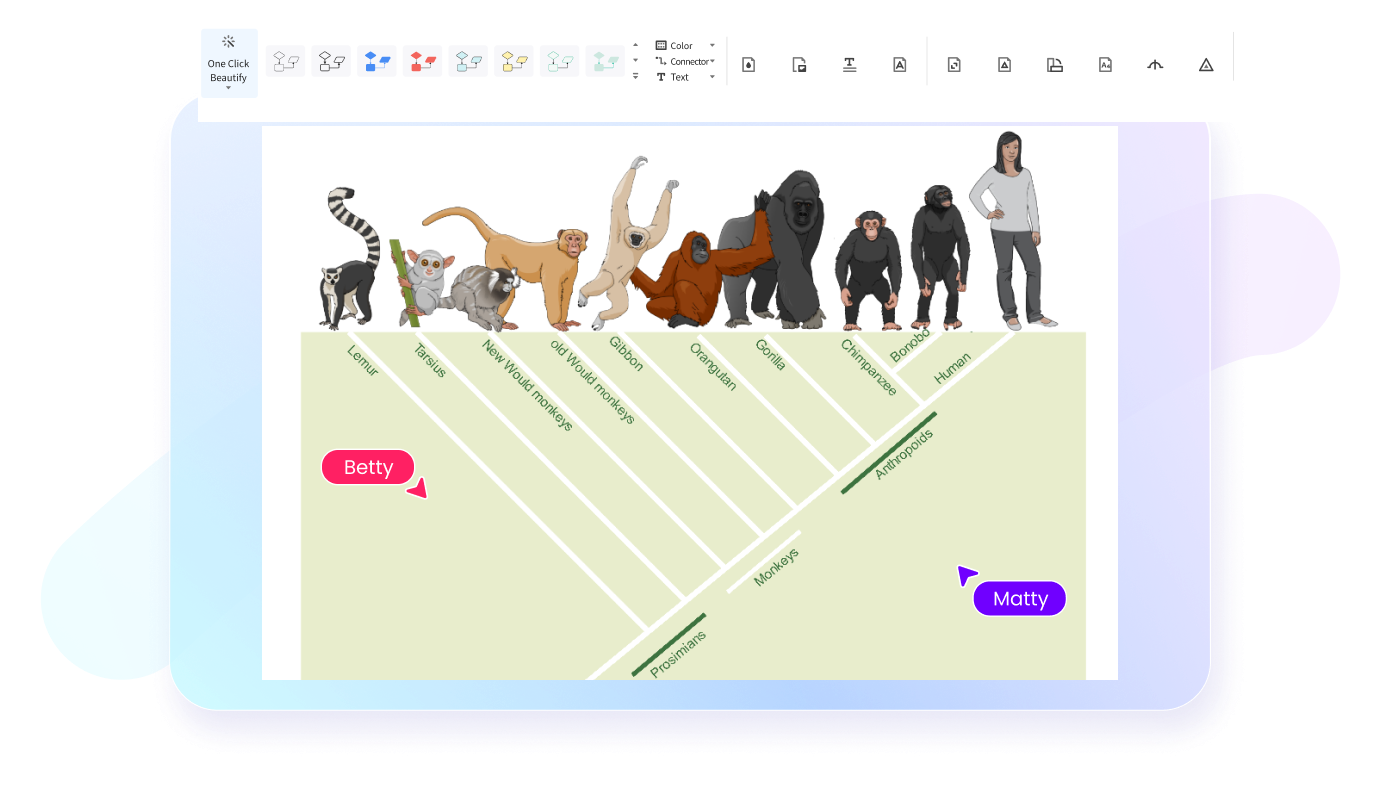
Phylogenetic tree software for everyone

Evolutionary Biologists
It helps them study the relationships between different species, understand patterns of evolution, and inferancestral relationships.

Taxonomists and Systematists
It helps in determining the placement of newly discovered species within existing taxonomic frameworks.

Non-profit Organizations
It aids in identifying orthologous and paralogous genes, studying gene family evolution, and inferring functional similarities anddifferences.
Why teams choose EdrawMax
Seamless Teamwork
EdrawMax facilitates real-time collaboration, allowing team members to work together on diagrams and share feedback instantly, fostering teamwork, and accelerating project progress.
Enhanced Efficency
With its intuitive interface, pre-made templates, and extensive symbol libraries, EdrawMax enables teams to create professional-quality diagrams quickly, saving time, reducing effort, and increasing productivity.
All in One
EdrawMax allows for making 280+ types of diagrams including flowcharts, mind mapps, Gantt charts, timelines and more, streamlining workflows and eliminating the need for multiple software, enhancing convenience.
Improved Communication
Use visually compelling diagrams to convey complex ideas, facilitating clear and concise communication within teams and with stakeholders, boosting understanding and decision-making processes.
Create phylogenetic trees with EdrawMax!
How to create a phylogenetic tree?
-
Data collection and alignment
 Gather the relevant geneticor morphological data for the organisms under study. This may involve sequencing DNA or collecting morphological measurements. Once the data is collected, it needs to be aligned to ensure that corresponding positions in the sequences or measurements are properly matched.
Gather the relevant geneticor morphological data for the organisms under study. This may involve sequencing DNA or collecting morphological measurements. Once the data is collected, it needs to be aligned to ensure that corresponding positions in the sequences or measurements are properly matched. -
Tree construction
 Construct the phylogenetic tree. There are several methods available for tree construction, including distance-based methods, parsimony methods, and likelihood-based methods. Each method has its own assumptions and algorithms. Researchers can choose the most appropriate methodbased on their data and research question.
Construct the phylogenetic tree. There are several methods available for tree construction, including distance-based methods, parsimony methods, and likelihood-based methods. Each method has its own assumptions and algorithms. Researchers can choose the most appropriate methodbased on their data and research question. -
Model selection and parameter estimation
 If likelihood-based methods are used, researchers need to selectan appropriate model of evolution that best fits their data. This involves choosing a substitution model that describes the rates and patterns of geneticor morphological changes. Parameter estimation is then performed to estimate the values of the model parameters.
If likelihood-based methods are used, researchers need to selectan appropriate model of evolution that best fits their data. This involves choosing a substitution model that describes the rates and patterns of geneticor morphological changes. Parameter estimation is then performed to estimate the values of the model parameters. -
Tree visualization and analysis
 Visualization tools allow researchers to customize the appearance of the tree, annotatebranches with additional information, and highlight specific clades or groups of interest. Statistical measures, such as bootstrap support or posteriorprobabilities, can be used to assess the robustness of the inferred relationships. Comparative analysis can be performed by mapping traits or other characteristicsonto the tree.
Visualization tools allow researchers to customize the appearance of the tree, annotatebranches with additional information, and highlight specific clades or groups of interest. Statistical measures, such as bootstrap support or posteriorprobabilities, can be used to assess the robustness of the inferred relationships. Comparative analysis can be performed by mapping traits or other characteristicsonto the tree.
What our users say
Explore more diagrams
FAQs about phylogenetic tree makers
-
Do I need prior knowledge of phylogenetics to use EdrawMax?While a basic understanding of phylogenetics could be helpful in accurately representing relationships, EdrawMax's user-friendly interface and templates allow you to create a phylogenetic tree even if you are not an expert in the field.Does EdrawMax have icons or symbols I can use for my phylogenetic tree?Yes. EdrawMax offers a library of icons, symbols, and shapes that you can use to enhance your phylogenetic tree. These include representations of organisms, genetic markers, and other relevant elements.Is there an automatic layout feature that arranges the tree for me?EdrawMax has an Edraw AI feature. It simplifies and optimizes diagrams, charts, and text creation and optimization. It can generate flowcharts, mind maps, and lists with a single command, perform copywriting with a single click, optimize diagram layout and style, and extract text from images using OCR.Can I collaborate with my team members?EdrawMax has collaboration features that enable you to work with team members in real time. These features allow multiple users to edit and comment on the same document simultaneously, making collaborating on phylogenetic tree projects easier. Just click the share button at the top right corner of your screen and invite your team.How can I flaunt my designs to others?One of the key features of EdrawMax is the ability to share and present. Through this integrated online feature, you can effortlessly showcase your diagrams as interactive slideshows. It also allows you to explain each aspect of your phylogenetic tree in a structured manner.














Types of phylogenetic trees